It shouldn’t surprise us that indeed in corridor of the genome where we do not obviously see a “ functional ” law( i.e., one that’s been evolutionarily fixed as a result of some picky advantage), there’s a type of law, but not like anything we ’ve preliminarily considered as similar. And what if it were doing commodity in three confines as well as the one dimension of the ATGC law? A paper just published in Bio Essays explores this tantalizing possibility
Is not it awful to have a really perplexing problem to gnaw on, one that generates nearly endless implicit explanations. How about “ what is all that non-coding DNA doing in genomes? ” — that98.5 of mortal inheritable material that does not produce proteins. To be fair, the deciphering ofnon-coding DNA is making great strides via the identification of sequences that are transcribed into RNAs that modulate gene expression, may be passed on transgenerationally( epigenetics) or set the gene expression program of a stem cell or specific towel cell. Massive quantities of reprise sequences( remnants of ancient retroviruses) have been set up in numerous genomes, and again, these do n’t law for protein, but at least there are believable models for what they ’re doing in evolutionary terms( ranging from genomic parasitism to symbiosis and indeed “ exploitation ” by the veritably host genome for producing the inheritable diversity on which elaboration works); apropos, somenon-coding DNA makes RNAs that silence these retroviral sequences, and retroviral accession into genomes is believed to have been the picky pressure for the elaboration of RNA hindrance( so– called RNAi); repetitious rudiments of colorful named types and tandem reprises pullulate; introns( numerous of which contain the forenamed types ofnon-coding sequences) have occurred to be pivotal in gene expression and regulation, most strikingly via indispensable splicing of the rendering parts that they separate.

Still, there’s plenitude of problem to gnaw on because although we’re decreasingly understanding the nature and origin of important of thenon-coding genome and are making major raids into its “ function ”( defined then as evolutionarily named, profitable effect on the host organism), we ’re far from explaining it all, and — further to the point — we ’re looking at it with a veritably low– exaggeration lens, so to speak. One of the interesting effects about DNA sequences is that a single sequence can “ render ” further than one piece of information depending on what’s “ reading ” it and in which direction – viral genomes are classic exemplifications in which genes read in one direction to produce a given protein imbrication with one or further genes read in the contrary direction( i.e., from the reciprocal beachfront of DNA) to produce different proteins. It’s a bit like making simple dispatches with rear– brace words( a so– called emordnilap). For illustration REEDSTOPSFLOW, which, by an imaginary reading device, could be divided into REED STOPS FLOW. Read backwards, it would give WOLF SPOTS DEER.
Now, if it’s of evolutionary advantage for two dispatches to be enciphered so economically – as is the case in viral genomes, which tend to evolve towards minimal complexity in terms of information content, hence reducing necessary coffers for reduplication — also the dispatches themselves evolve with a high degree of constraint. What does this mean? Well, we could word our original illustration communication as RUSH– STEM IMPEDES CURRENT, which would embody the same essential information as REED STOPS FLOW. still, that communication, if read in rear( or indeed in the same sense, but in different gobbets) doesn’t render anything fresh that’s particularly meaningful. presumably the only way of conveying both pieces of information in the original dispatches contemporaneously is the very wording REEDSTOPSFLOW that’s a largely constrained system! Indeed, if we studied enough exemplifications of rear– brace expressions in English, we’d see that they are, on the whole, made up of rather short words, and the sequences are missing certain units of language similar as papers( the, a); if we looked more nearly, we might indeed descry a lesser representation than normal of certain letters of the ABC in similar dispatches. We’d see these as impulses in word and letter operation that would, a priori, allow us to have a stab at relating similar “ binary– function ” pieces of information.

Now let’s return to the “ letters ”, “ words ”, and “ information ” decoded in genomes. For two distinct pieces of information to be decoded in the same piece of inheritable sequence we would, also, anticipate the constraints to be manifest in impulses of word and letter operation — the circumlocutions, independently, for amino acid sequences constituting proteins, and their three- letter law. Hence a sequence of DNA can decode for a protein and, in addition, for commodity differently. This “ commodity additional ”, according to Giorgio Bernardi, is information that directs the packaging of the enormous length of DNA in a cell into the fairly bitsy nexus. Primarily it’s the law that guides the list of the DNA- packaging proteins known as histones. Bernardi refers to this as the “ genomic law ” — a structural law that defines the shape and contraction of DNA into the largely– condensed form known as “ chromatin ”.
But did n’t we start with an explanation fornon-coding DNA, not protein- rendering sequences? Yes, and in the long stretches ofnon-coding DNA we see information in excess of bare reprises, tandem reprises and remnants of ancient retroviruses there’s a type of law at the position of preference for the GC brace of chemical DNA bases compared with AT. As Bernardi reviews, synthesizing his and others ’ groundbreaking work, in the core sequences of the eukaryotic genome, the GC content in structural organizational units of the genome nominated “ isochores ” increased during the evolutionary transition between so– called cold– thoroughbred and warm– thoroughbred organisms. And, fascinatingly, this sequence bias overlaps with sequences that are much more constrained in function these are the veritably protein- rendering sequences mentioned before, and they — further than the intermediatingnon-coding sequences are the indication to the “ genomic law ”.
Protein- rendering sequences are also packed and condensed in the nexus – particularly when they ’re not “ in use ”( i.e., being transcribed, and also restated into protein) – but they also contain fairly constant information on precise amino acid individualities, else they would fail to render proteins rightly elaboration would act on similar mutations in a largely negative manner, making them extremely doubtful to persist and be visible to us. But the amino acid law in DNA has a little “ catch ” that evolved in the most simple of unicellular organisms( bacteria and archaea) billions of times ago the law is incompletely spare. For illustration, the amino acid Threonine can be enciphered in eukaryotic DNA in no smaller than four ways ACT, ACC, ACA or ACG. The third letter is variable and hence “ available ” for the coding of redundant information. This is exactly what happens to produce the “ genomic law ”, in this case creating a bias for the ACC and ACG forms in warm– thoroughbred organisms. Hence, the high constraint on this fresh “ law ” — which is also seen in corridor of the genome that aren’t under similar constraint as protein- rendering sequences is assessed by the packaging of protein- rendering sequences that embody two sets of information contemporaneously. This is similar to our illustration of the largely– constrained binary– information sequence REEDSTOPSFLOW.
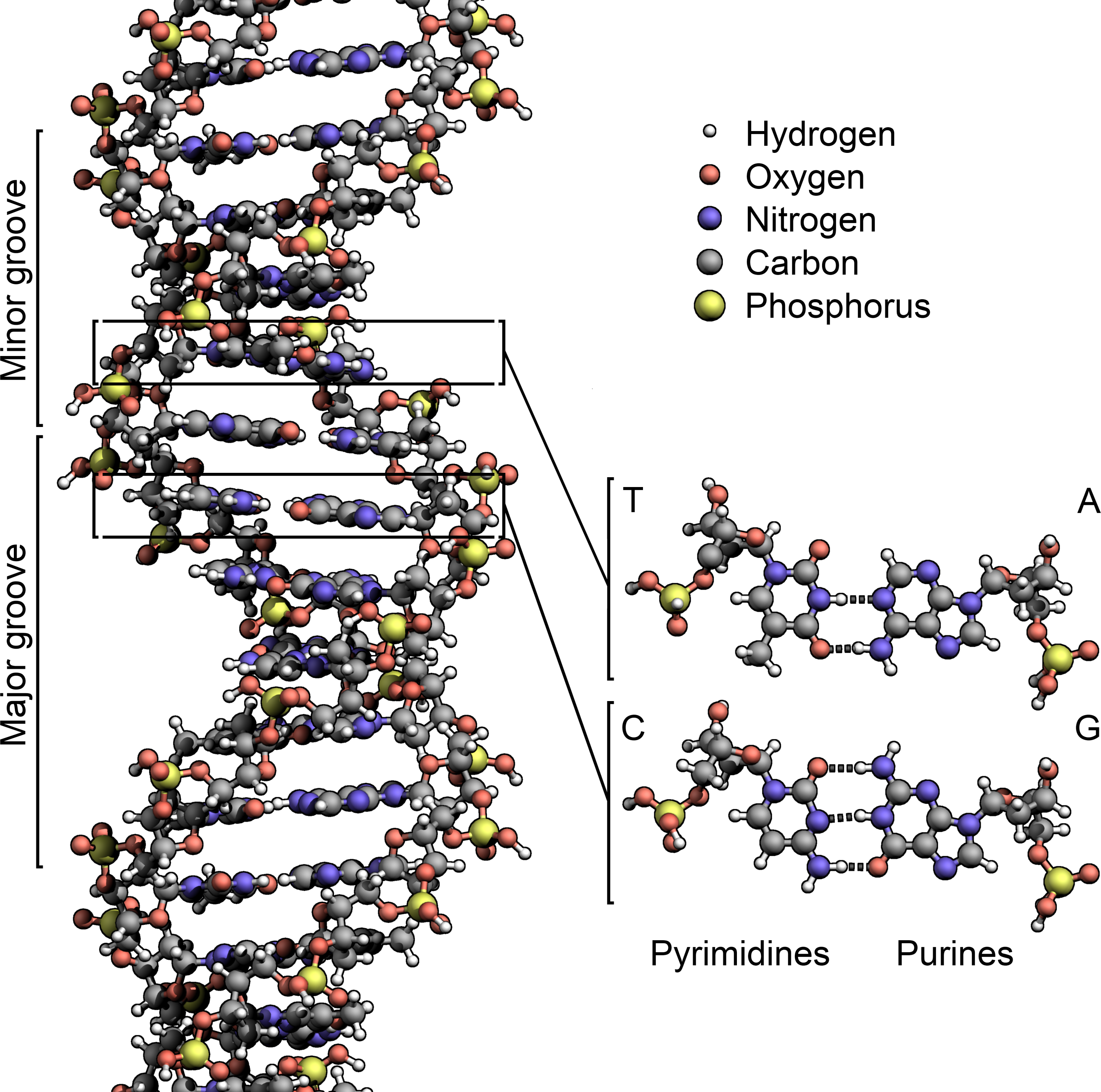
Importantly, still, the constraint isn’t as strict as in our English language illustration because of the redundancy of the third position of the triplet law for amino acids a better analogy would be SHE * ATE * STU * where the asterisk stands for a variable letter that does not make any difference to the machine that reads the three- letter element of the four- letter communication. One could also imagine a alternate position of information formed by adding “ D ” at these asterisk points, to make SHEDATEDSTUD( SHE DATED superstud).
Next imagine a alternate reading machine that looks for meaningful expressions of a “ sensitive nature ” containing a lesser than average attention of Ds. This reading machine carries a folding machine with it that places a kind of cut at each D, kinking the communication by 120 degrees in a aeroplane . a point where the communication should be bent by 120 degrees in the same aeroplane , we’d end up with a more compact, triangular, interpretation. In eukaryotic genomes, the GC sequence bias proposed to be responsible for structural condensation extends into non-coding sequences, some of which have linked conditioning, though less constrained in sequence than protein- rendering DNA. There it directs their condensation via histone- containing nucleosomes to form chromatin.
These regions of DNA may also be regarded as structurally important rudiments in forming the correct shape and separation of condensed coding sequences in the genome, anyhow of any other possible function that those non-coding sequences have in substance, this would be an “ explanation ” for the continuity in genomes of sequences to which no “ function ”( in terms of evolutionarily- named exertion), can be credited( or, at least, no substantial function).
A final analogy this time much more nearly affiliated — might be the veritably amino acid sequences in large proteins, which do a variety of twists, turns, crowds etc. We may marvel at similar complicated structures and ask “ but do they need to be relatively so complicated for their function? ” Well, perhaps they do in order to condense and place corridor of the protein in the exact exposure and place that generates the three- dimensional structure that has been successfully named by elaboration. But with a knowledge that the “ genomic law ” overlaps protein rendering sequences, we might indeed start to come suspicious that there’s another picky pressure at work as well.